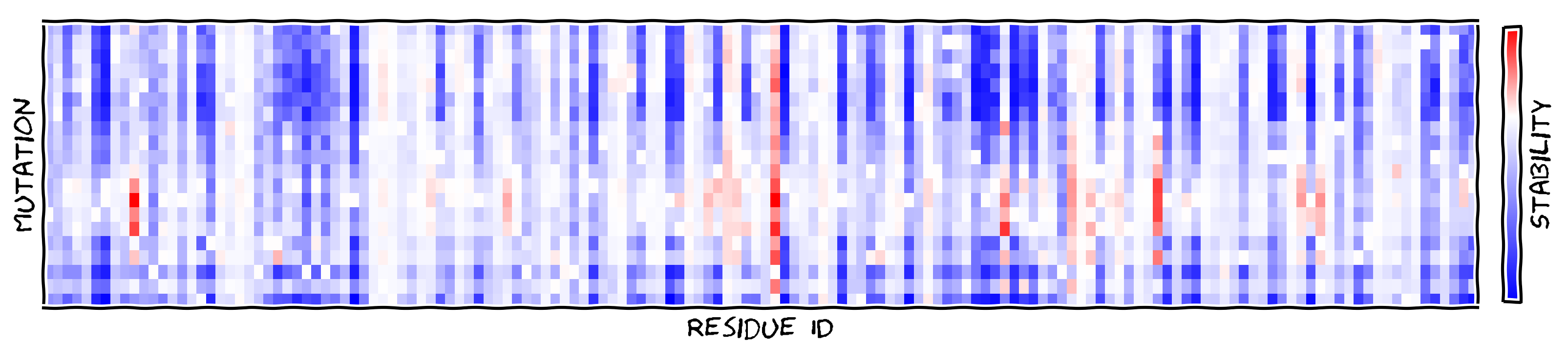
The DeepDDG server predicts the stability change of protein point mutations using neural networks.
The dataset used to train the network can be downloaded here.
For a quick demo, use the T4 lysozyme structure and the predicted result is like this.